Configuration templates¶
Here we present some configuration templates that we used during our experiments. Used on the .tif sequence above, they produce good results. In order for you to see what we call “good” results, they are shown above too for each configuration template.
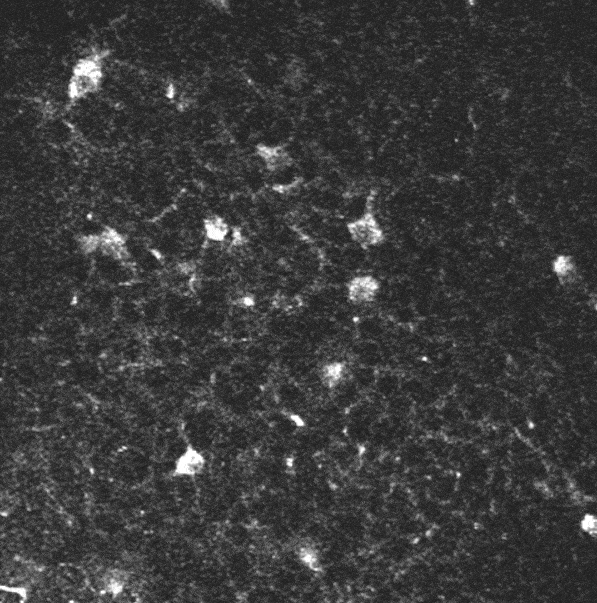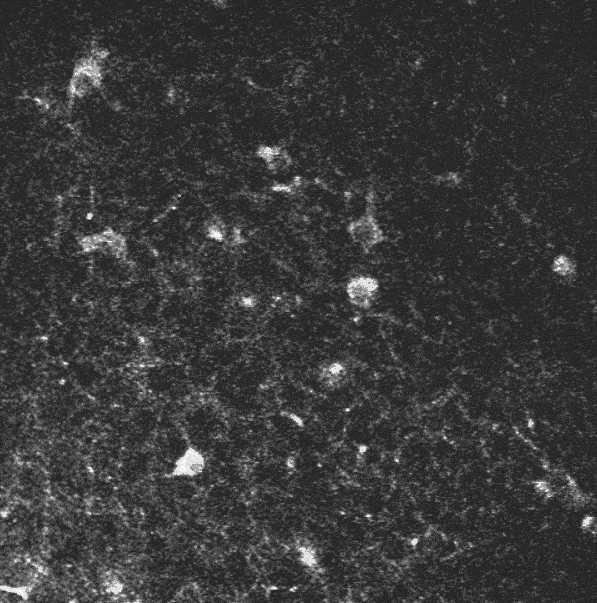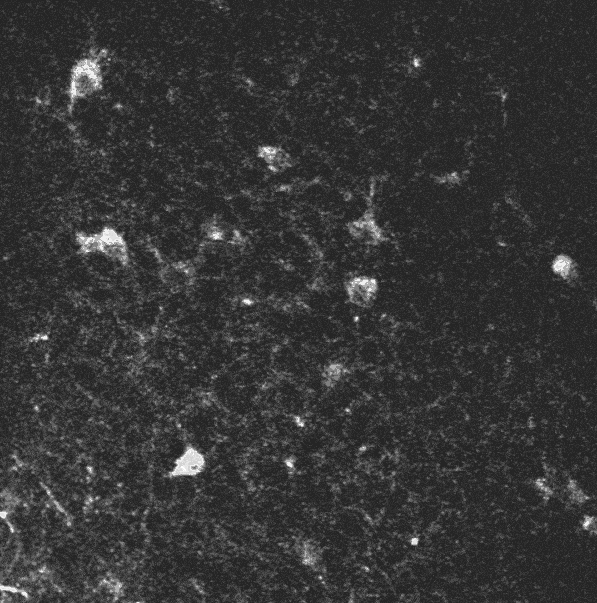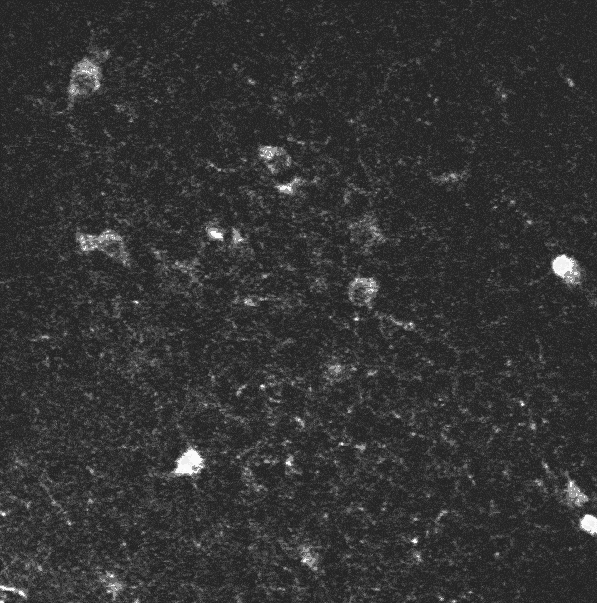
The initial sequence.
The configuration templates can be found in the pyamnesia/config_templates/ directory.
Clustering templates¶
Skeleton pixels clustering¶
- file:
config_clustering_pixel.yaml - context: clustering the traces of skeleton pixels from the
.tifsequence. - specs:
projection_substructureis set to'skeleton'andprojection_methodto'pixel'. t-SNE perplexity is relatively high, because we expect to find mesoscopic clusters composed of dozens of pixels. - results:
Clustering plot, cluster overlay and cluster traces.
Skeleton branches clustering¶
- file:
config_clustering_branch.yaml - context: clustering the traces of skeleton branches (or mesoscopic clusters from the pixel clustering) from the
.tifsequence. - specs:
projection_substructureis set to'skeleton'andprojection_methodto'branch'. t-SNE perplexity is lower, because we expect to find macroscopic clusters composed of a few branches (or mesoscopic clusters). - results:
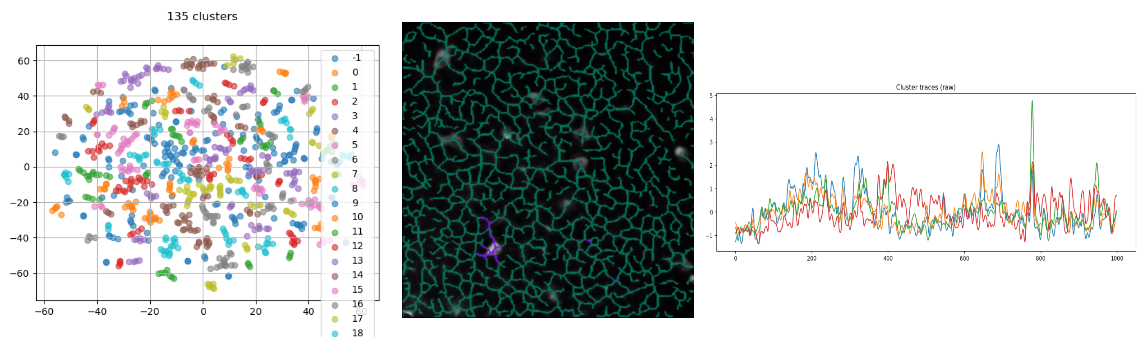
Clustering plot, cluster overlay and cluster traces.
Active pixels clustering¶
- file:
config_clustering_active.yaml - context: clustering the traces of active pixels from the
.tifsequence. - specs:
projection_substructureis set to'active'andprojection_methodto'pixel'. t-SNE perplexity is lower, because we expect to find macroscopic clusters composed of a few branches (or mesoscopic clusters). - results:
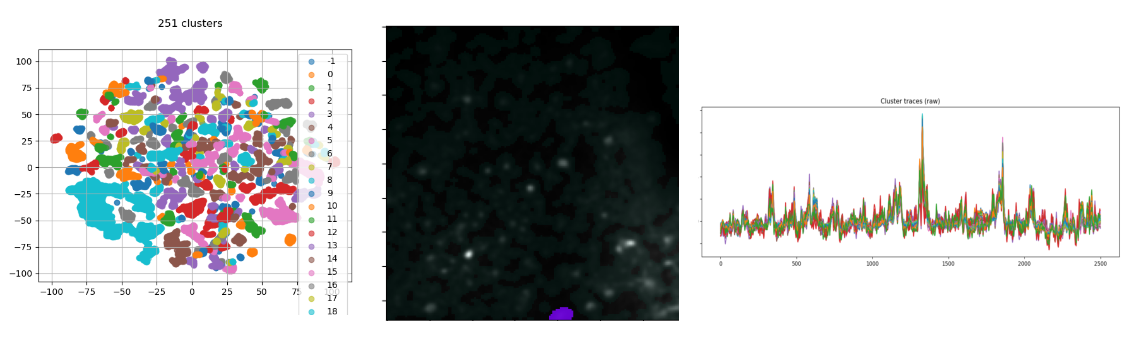
Clustering plot, cluster overlay and cluster traces.
Factorization templates¶
NMF¶
- file:
config_factorization_nmf.yaml - context: performing a NMF on the
.tifsequence. - specs:
element_normalization_methodis set tonullso that the input of the NMF is nonnegative. The number of components is set to50but can be adapted. - results:
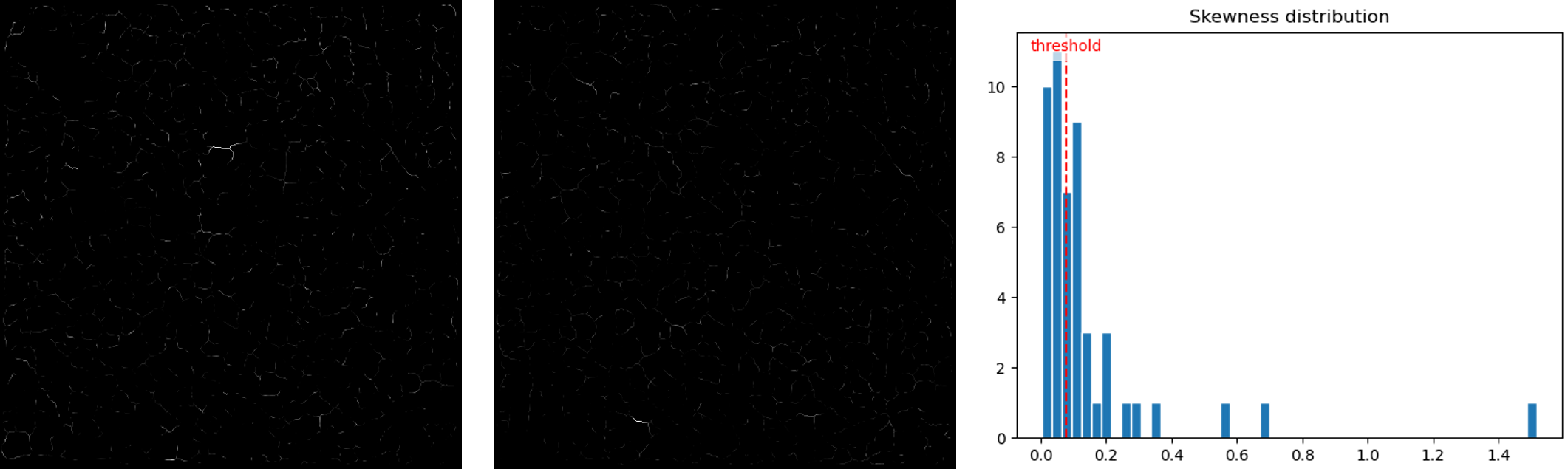
Two components and the skewness histogram (open image in new tab for a better view).
PCA and ICA¶
- file:
config_factorization_pca_ica.yaml - context: performing a PCA and an ICA on the
.tifsequence. - specs:
element_normalization_methodis set to'z-score'. The number of components is set to50but can be adapted. - results:
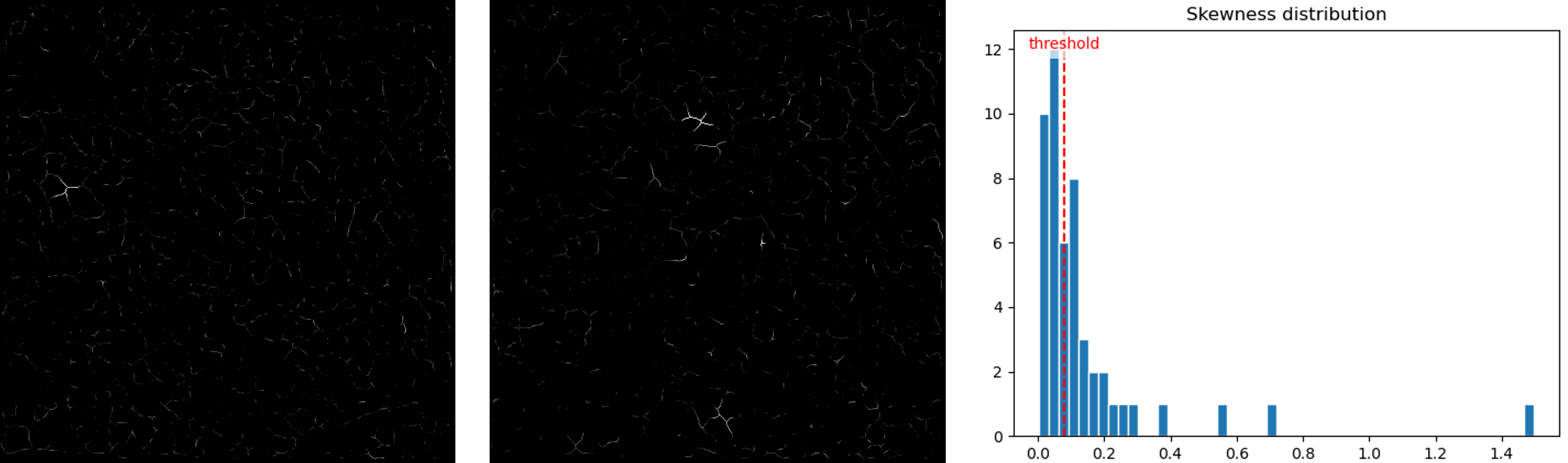
Two components and the skewness histogram (open image in new tab for a better view).